Genomic prediction and design now require models that connect local motifs with megabase scale regulatory context and that operate across many organisms. Nucleotide Transformer v3, or NTv3, is InstaDeep’s new multi species genomics foundation model for this setting. It unifies representation learning, functional track and genome annotation prediction, and controllable sequence generation in a single backbone that runs on 1 Mb contexts at single nucleotide resolution.
Earlier Nucleotide Transformer models already showed that self supervised pretraining on thousands of genomes yields strong features for molecular phenotype prediction. The original series included models from 50M to 2.5B parameters trained on 3,200 human genomes and 850 additional genomes from diverse species. NTv3 keeps this sequence only pretraining idea but extends it to longer contexts and adds explicit functional supervision and a generative mode.
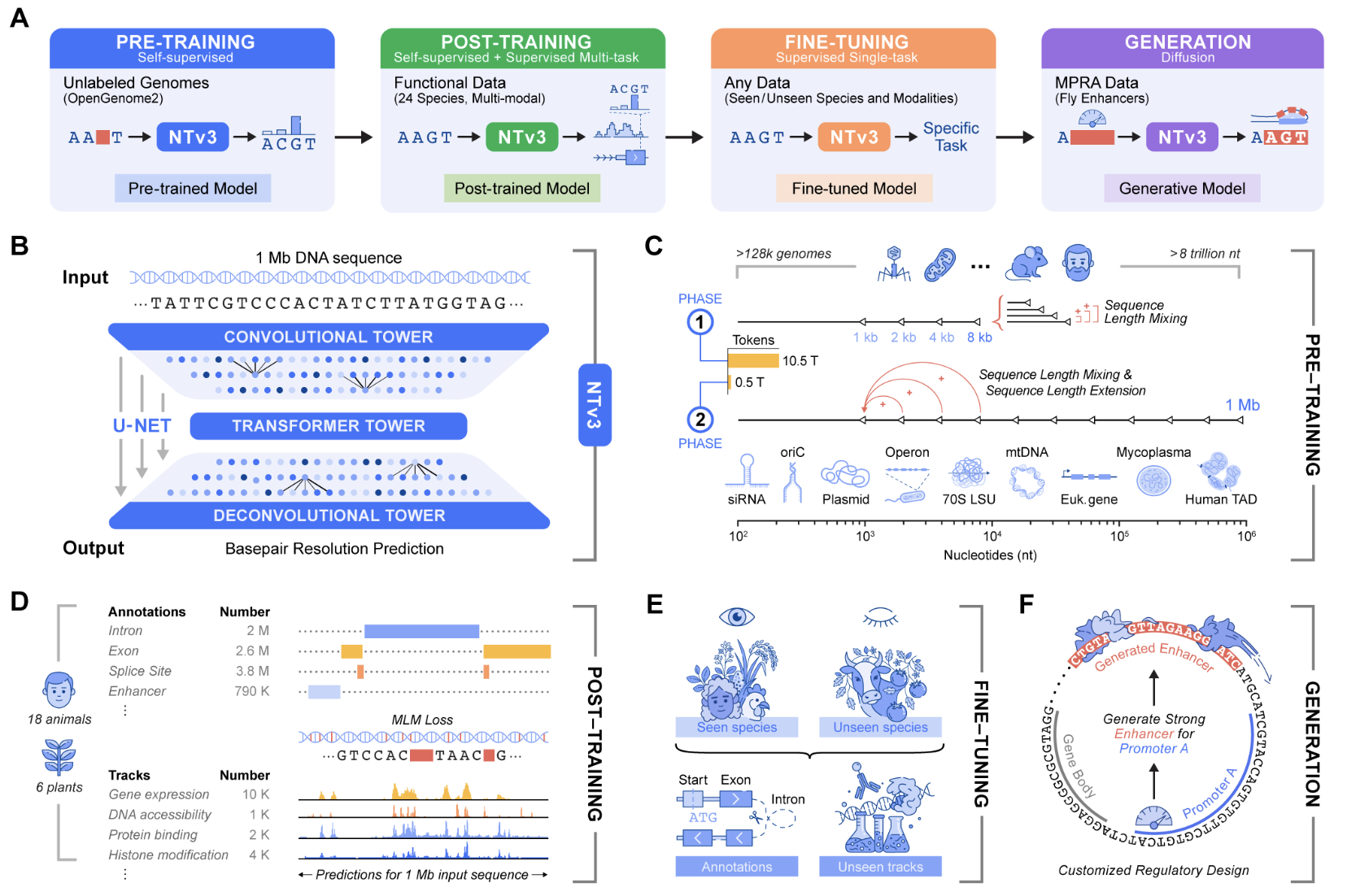
Architecture for 1 Mb genomic windows
NTv3 uses a U-Net style architecture that targets very long genomic windows. A convolutional downsampling tower compresses the input sequence, a transformer stack models long range dependencies in that compressed space, and a deconvolution tower restores base level resolution for prediction and generation. Inputs are tokenized at the character level over A, T, C, G, N with special tokens such as , , , , , and . Sequence length must be a multiple of 128 tokens, and the reference implementation uses padding to enforce this constraint. All public checkpoints use single base tokenization with a vocabulary size of 11 tokens.
The smallest public model, NTv3 8M pre, has about 7.69M parameters with hidden dimension 256, FFN dimension 1,024, 2 transformer layers, 8 attention heads, and 7 downsample stages. At the high end, NTv3 650M uses hidden dimension 1,536, FFN dimension 6,144, 12 transformer layers, 24 attention heads, and 7 downsample stages, and adds conditioning layers for species specific prediction heads.
Training data
The NTv3 model is pretrained on 9 trillion base pairs from the OpenGenome2 resource using base resolution masked language modeling. After this stage, the model is post trained with a joint objective that integrates continued self supervision with supervised learning on approximately 16,000 functional tracks and annotation labels from 24 animal and plant species.
Performance and Ntv3 Benchmark
After post training NTv3 achieves state of the art accuracy for functional track prediction and genome annotation across species. It outperforms strong sequence to function models and previous genomic foundation models on existing public benchmarks and on the new Ntv3 Benchmark, which is defined as a controlled downstream fine tuning suite with standardized 32 kb input windows and base resolution outputs.
The Ntv3 Benchmark currently consists of 106 long range, single nucleotide, cross assay, cross species tasks. Because NTv3 sees thousands of tracks across 24 species during post training, the model learns a shared regulatory grammar that transfers between organisms and assays and supports coherent long range genome to function inference.
From prediction to controllable sequence generation
Beyond prediction, NTv3 can be fine tuned into a controllable generative model via masked diffusion language modeling. In this mode the model receives conditioning signals that encode desired enhancer activity levels and promoter selectivity, and it fills masked spans in the DNA sequence in a way that is consistent with those conditions.
In experiments described in the launch materials, the team designs 1,000 enhancer sequences with specified activity and promoter specificity and validates them in vitro using STARR seq assays in collaboration with the Stark Lab. The results show that these generated enhancers recover the intended ordering of activity levels and reach more than 2 times improved promoter specificity compared with baselines.
Comparison Table
| Dimension | NTv3 (Nucleotide Transformer v3) | GENA-LM |
|---|---|---|
| Primary goal | Unified multi species genomics foundation model for representation learning, sequence to function prediction and controllable sequence generation | Family of DNA language models for long sequences focused on transfer learning for many supervised genomic prediction tasks |
| Architecture | U-Net style convolutional tower, transformer stack, deconvolutional tower, single base resolution language model, post trained versions add multi species conditioning and task specific heads | BERT based encoder models with 12 or 24 layers and BigBird variants with sparse attention, extended further with recurrent memory transformer for long contexts |
| Parameter scale | Family spans 8M, 100M and 650M parameters | Base models have 110M parameters and large models have 336M parameters, including BigBird variants at 110M |
| Native context length | Up to 1 Mb input at single nucleotide resolution for both pre trained and post trained models | Up to about 4500 bp with 512 BPE tokens for BERT models and up to 36000 bp with 4096 tokens for BigBird models |
| Extended context mechanism | Uses U-Net style convolutional tower to aggregate long range context before transformer layers while keeping single base resolution; context length is fixed at 1 Mb in the released checkpoints | Uses sparse attention in BigBird variants plus recurrent memory transformer to extend effective context to hundreds of thousands of base pairs |
| Tokenization | Character level tokenizer over A, T, C, G, N and special tokens; each nucleotide is a token | BPE tokenizer on DNA that maps to about 4500 bp for 512 tokens; two tokenizers are used, one on T2T only and one on T2T plus 1000G SNPs plus multispecies data |
| Pretraining corpus size | First stage pre training on OpenGenome2 with about 9 trillion base pairs from more than 128000 species | Human only models trained on pre processed human T2T v2 plus 1000 Genomes SNPs, about 480 × 10^9 base pairs, multispecies models trained on combined human and multispecies data, about 1072 × 10^9 base pairs |
| Species coverage | More than 128000 species in OpenGenome2 pretraining and post training supervision from 24 animal and plant species | Human focused models plus taxon specific models for yeast, Arabidopsis and Drosophila and multispecies models from ENSEMBL genomes |
| Supervised post training signals | About 16000 functional tracks across about 10 assay types and about 2700 tissues in 24 species, used to condition the backbone with discrete labels and to train functional heads | Fine tuned on multiple supervised tasks, including promoters, splice sites, Drosophila enhancers, chromatin profiles and polyadenylation sites, with task specific heads on top of the LM |
| Generative capabilities | Can be fine tuned into a controllable generative model using masked diffusion language modeling, used to design 1000 promoter specific enhancers that achieved more than 2× increased specificity in STARR seq assays | Primarily used as a masked language model and feature extractor, supports sequence completion through MLM but the main publication focuses on predictive tasks rather than explicit controllable sequence design |
Key Takeaways
- NTv3 is a long range, multi species genomics foundation model: It unifies representation learning, functional track prediction, genome annotation, and controllable sequence generation in a single U Net style architecture that supports 1 Mb nucleotide resolution context across 24 animal and plant species.
- The model is trained on 9 trillion base pairs with joint self supervised and supervised objectives: NTv3 is pretrained on 9 trillion base pairs from OpenGenome2 with base resolution masked language modeling, then post trained on more than 16,000 functional tracks and annotation labels from 24 species using a joint objective that mixes continued self supervision with supervised learning.
- NTv3 achieves state of the art performance on the Ntv3 Benchmark: After post training, NTv3 reaches state of the art accuracy for functional track prediction and genome annotation across species and outperforms previous sequence to function models and genomics foundation models on public benchmarks and on the Ntv3 Benchmark, which contains 106 standardized long range downstream tasks with 32 kb input and base resolution outputs.
- The same backbone supports controllable enhancer design validated with STARR seq: NTv3 can be fine tuned as a controllable generative model using masked diffusion language modeling to design enhancer sequences with specified activity levels and promoter selectivity, and these designs are validated experimentally with STARR seq assays that confirm the intended activity ordering and improved promoter specificity.
Check out theRepo,Model on HFandTechnical details.Also,feel free to follow us onTwitterand don’t forget to join our100k+ ML SubRedditand Subscribe toour Newsletter. Wait! are you on telegram?now you can join us on telegram as well.
Asif Razzaq is the CEO of Marktechpost Media Inc.. As a visionary entrepreneur and engineer, Asif is committed to harnessing the potential of Artificial Intelligence for social good. His most recent endeavor is the launch of an Artificial Intelligence Media Platform, Marktechpost, which stands out for its in-depth coverage of machine learning and deep learning news that is both technically sound and easily understandable by a wide audience. The platform boasts of over 2 million monthly views, illustrating its popularity among audiences.